The molecular machinery used by ‘superbugs’ to resist chemicals designed to kill them could also help produce precursors for a new generation of nylon and other polymers, according to new research.
Scientists from the University of Leeds have collaborated with Australian researchers to identify ancient protein pumps that make infectious bacteria tough to treat, but which could be key to developing new environmentally-friendly polymers.
Bacteria, called ‘superbugs’, that are unaffected by antiseptics and antibiotics are a growing problem, but exactly how they develop resistance is often not fully understood.
Previous research by the University of Leeds and Macquarie University, Australia, revealed how a bacterium called Acinetobacter baumannii resisted chlorhexidine, a powerful hospital-grade antiseptic listed by the World Health Organisation as an “essential medicine”.
A. baumannii’s secret weapon, they found, is a protein called AceI, which sits on its surface and pumps out any chlorhexidine that gets inside. That was a surprise to researchers, because the protein has existed for millions of years longer than the antiseptic.
“It may still be years away, but producing plastic using bacteria would dramatically reduce its carbon footprint, as it would avoid the need to use the petrochemicals that we rely on today.”
“Resistance to artificial antiseptics appears to be a lucky accident for the bacteria, and it could also be useful for humans,” says Professor Ian Paulsen from Australia’s Macquarie University, one of the leaders of the research group.
“The gene that encodes the AceI protein appears to be very old, but chlorhexidine was only created in the twentieth century,” says lead author of the new research, Dr Karl Hassan, who carried out key elements of the work at the University of Leeds and is now at Australia’s University of Newcastle.
“So the gene can’t have the native function of protecting against chlorhexidine. It’s a side reaction that is fortunate for the bacteria.”
Now the research team have looked at what other compounds are transported by AceI and its relations, collectively known as Proteobacterial Antimicrobial Compound Efflux (PACE) proteins.
They found good news and bad news. The bad news was that PACE proteins are likely to be future engines of antimicrobial resistance. The good news is that their ability to transport a wide range of substances means that they might be effectively repurposed in an industrial context to catalyse the manufacture of “petroleum-free” polymers such as nylons.
Furthermore, this new understanding of the activities of these transport proteins means researchers can try to develop inhibitors which stop them overcoming the antibiotics.
“These PACE proteins, which we have isolated and purified in Leeds, are promiscuous in the compounds that they transport and are a likely cause of future resistance to new antimicrobials that are currently being developed,” says co-author Professor Peter Henderson from the University of Leeds’ School of Biomedical Sciences.
The PACE proteins may have applications in green biotechnology, as the naturally occurring molecules they transport are a potential alternative to petroleum as the basis of polymers such as nylon.
Professor Henderson, who is also part of the University's Astbury Centre, continued: “We’ve shown in this study that AceI can transport a substance called cadaverine out of the cell, and that is already used in the manufacture of plastics today.
“It may still be years away, but producing plastic using bacteria would dramatically reduce its carbon footprint, as it would avoid the need to use the petrochemicals that we rely on today.”
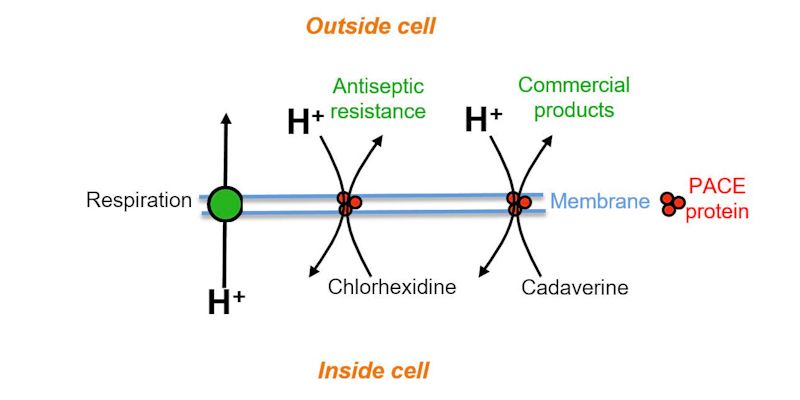
Figure shows how the PACE protein pump transports antiseptic - chlorhexidine - and a plastic precursor - cadaverine - out of the cell
Researchers in Leeds are exploring the second potential application of this research, attempting to develop an inhibitor of these protein pumps. They hope that by stopping the protein pump from working they could provide a method of killing ‘superbugs’ that are otherwise resistant to antibiotics, to help tackle this growing global health threat.
The research was published in the Proceedings of the National Academy of Sciences, and was funded by the European Commission and the National Health and Medical Research Council of Australia.
Further information
For interview requests please contact Simon Moore, Press Officer at the University of Leeds, on 0113 34 38059 or s.i.moore@leeds.ac.uk.
The paper, published in the journal Proceedings of the National Academy of Sciences, is titled ‘Short-chain diamines are the physiological substrates of PACE family efflux pumps’ and is available online here: www.pnas.org/cgi/doi/10.1073/pnas.1901591116